OnePot cDNA和gDNA文库制备试剂盒 cDNA & gDNA Library Prep Kit
产品说明书
FAQ
COA
已发表文献
Hieff NGS® OnePot cDNA & gDNA Library Prep Kit是针对Illumina®或者MGI®测序平台专业开发设计的新一代酶切法建库试剂盒。与传统的建库法比较,本品采用高质量的片段化酶,摆脱了繁琐的超声过程,同时简化了操作流程,将片段化模块与末端修复模块合二为一,极大的降低了建库的时间和成本。本试剂盒具有优秀的文库转化率,可应用于常规动植物基因组、微生物基因组等样本,同时能兼容cfDNA样本的建库。该试剂盒使用了最新优化的连接酶,改善了接头连接时的片段自连现象,同时替换了新型高保真酶,进一步提升了扩增的均一性和保真性。
Ø 适用500 pg-1 μg的基因组DNA、全长cDNA(衔接Hieff NGS® ds-cDNA Synthesis Kit全长cDNA合成试剂盒(Yeasen Cat#13488))等样本;
Ø 高质量片段化酶,可随机切割双链DNA,酶切片段无偏好性;
Ø 片段化、末端修复/加A一步完成;
Ø 强扩增效率的高保真酶,显著提高文库质量及产量;
Ø 适用于cfDNA样本;
Ø 严格的批次性能与稳定性质控;
产品组分
|
组分编号 组分名称 |
13502ES24 |
13502ES96 |
||
|
13502-A |
 |
Smearase Buffer |
240 μL |
960 μL |
|
13502-B |
 |
DNA Extra-working Buffer |
240 μL |
960 μL |
|
13502-C |
 |
Smearase Enzyme Mix |
120 μL |
480 μL |
|
13502-D |
 |
Ligation Enhancer |
720 μL |
2×1440 μL |
|
13502-E |
 |
Novel T4 DNA Ligase |
120 μL |
480 μL |
|
13502-F |
 |
2× Ultima HF Amplification Mix |
600 μL |
2×1200 μL |
运输与保存方法
干冰运输。-20℃保存。有效期1年。
注意事项
一、关于操作
1. 为了您的安全和健康,请穿实验服并戴一次性手套操作。
2. 请于使用前将试剂盒各组分置于室温解冻。解冻后上下颠倒数次充分混匀,短暂离心后置于冰上待用。
3. 配制各步骤反应液时推荐使用移液器吹打混匀或轻轻振荡,剧烈振荡可能会造成文库产出下降。
4. 为避免样品交叉污染,推荐使用带滤芯的枪头,吸取不同样品时请更换枪头。
5. 推荐在带热盖的PCR仪中进行各步骤反应,使用前应预热PCR仪至反应温度附近。
6. PCR产物因操作不当极容易产生气溶胶污染,进而影响实验结果准确性。推荐将PCR反应体系配制区和PCR产物纯化检测区进行强制性的物理隔离;使用专用的移液器等设备;并定时对各实验区域进行清洁(使用0.5%次氯酸钠或10%漂白剂进行擦拭清理),以保证实验环境的洁净度。
7. 本产品仅作科研用途!
二、关于接头连接(Adapter Ligation)
Illumina接头:
1. 本公司可提供短接头(也称为小Y接头、不完整接头)试剂盒,客户可根据实验需求进行选择。
目前有双端 384 种 Index Primers: Hieff NGS® 384 CDI Primer for Illumina® , Set 1~Set 2 (Cat#12412~Cat#12413)。
2. 我们建议选用高质量的商业化接头。如客户使用自制接头,请委托具有NGS引物合成经验的公司,并备注需进行严格的防污染控制。此外,进行接头退火操作时,请在超净台完成。每次只操作一种接头,防止交叉污染。
3. 使用接头时,请提前将接头取出放在4°C或冰盒上解冻;在室温操作时,实验室温度最好不要超过25°C,防止接头解链。
4. 建库过程中,接头浓度过高或过低都会导致建库成功率变低。本试剂盒操作方案中,所加入的接头体积固定为5 μL,请根据初始的DNA或者RNA投入量,参考表1对接头进行稀释。本公司接头原始浓度均为15 μM,接头稀释液请选择0.1×TE buffer,稀释过的接头可在4°C保存48小时。
表1 Input Total RNA/DNA量与接头使用浓度推荐表
|
Input Total RNA(参照Cat#13488 RNA投入量) |
Adapter stock concentration |
Input Total DNA |
Adapter stock concentration |
|
≥10 ng |
15 μM |
1μg~200 ng |
15 μM |
|
<10 ng |
3 μM |
100 ng |
10 μM |
|
50 ng |
5 μM |
||
|
10 ng |
3 μM |
||
|
5 ng |
1μM |
||
|
≤1ng |
0.5μM |
三、关于文库扩增(Library Amplification)
文库扩增步骤需要严格控制扩增循环数。循环数不足,将导致文库产量低;循环数过多,又将导致文库偏好性增加、重复度增加、嵌合产物增加、扩增突变积累等多种不良后果。表2列举了使用本试剂盒,获得1mg文库的推荐循环数。
表 2 Input Total RNA或者DNA量与扩增循环数推荐表*
|
Input Total RNA |
Number of cycles |
Input Total DNA |
Number of cycles |
|
<1 ng |
10~12 |
<1 ng |
14~16 |
|
1ng |
9~10 |
1 ng |
13~14 |
|
10 ng |
6~7 |
5 ng |
10~11 |
|
50ng |
4~5 |
10 ng |
9~10 |
|
100~1000 ng |
4 |
50 ng |
7~8 |
|
100 ng |
6~7 |
||
|
200 ng |
5~6 |
【注】:*由于文库产量不仅与投入量和扩增循环数相关,样本质量等都会影响产量。建库过程中请根据实际情况综合考虑,选择最合适的建库条件。
四、DNA磁珠纯化与分选(Bead-based Clean Up and Size Selection)
1. 建库过程中有多个步骤需要使用DNA纯化磁珠,我们推荐使用Hieff NGS® DNA Selection Beads (Yeasen Cat#12601)或AMPure® XP磁珠(Beckman Cat#A63880)进行DNA纯化和分选。
2. 磁珠使用前应先平衡至室温,否则会导致得率下降、分选效果不佳。
3. 磁珠每次使用前都应充分振荡混匀或使用移液器上下吹打充分混匀。
4. 转移上清时,请勿吸取磁珠,即使微量残留都将影响后续文库质量。
5. 磁珠洗涤使用的80%乙醇应现用现配,否则将影响回收效率。
6. 产物洗脱前应将磁珠置于室温干燥。干燥不充分容易造成无水乙醇残留影响后续反应;过分干燥又会导致磁珠开裂进而降低纯化得率。通常情况下,室温干燥3-5 min足以让磁珠充分干燥。
7. DNA纯化或长度分选产物如需保存,可使用0.1×TE Buffer洗脱,产物于4°C可保存2天,-20°C可保存1个月。
五、关于文库质检(Library Quality Analysis)
1. 通常情况下,构建好的文库可通过长度分布检测和浓度检测来进行质量评价。
2. 文库浓度检测可使用:基于双链DNA荧光染料的方法,如Qubit®、PicoGreen®等;基于qPCR绝对定量的方法。
3. 推荐使用qPCR方法进行文库浓度检测:Qubit®等基于双链DNA荧光染料的浓度测定方法时,无法有效区分单端连接Adapter的产物、两端均未连接Adapter的产物及其他不完整双链结构产物;qPCR绝对定量基于PCR扩增原理,仅定量样品中两端Adapter完整的文库(即可测序的文库),可排除单端或双端都不连接Adapter的不可测序文库的干扰。
4. 文库浓度检测不可使用:基于光谱检测的方法,如NanoDrop®等。
5. 文库长度分布检测,可通过Agilent Bioanalyzer 2100等基于毛细管电泳或微控流原理的设备进行检测。
六、自备材料(Other Material)
1. DNA纯化磁珠:Hieff NGS® DNA Selection Beads(Yeasen Cat#12601)或AMPure® XP Beads(A63880)或其他等效产品。
2. Adapters:含Index的长接头(Yeasen Cat#12615~12618)或者无Index的短接头试剂盒(Yeasen Cat#12611~12614)。
3. 文库质检:Agilent 2100 Bioanalyzer DNA 1000 Chip/ High Sensitivity Chip或其他等效产品;文库定量试剂。
4. 其他材料:无水乙醇、无菌超纯水、低吸附枪头、PCR管、磁力架、PCR仪等。
建库流程图
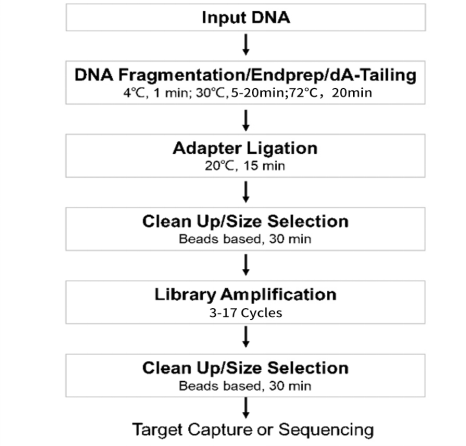
图1 DNA建库操作流程
使用方法
Step 1 cDNA & gDNA片段化/末端修复/dA尾添加(DNA Fragment/End Preparation/dA-Tailing)
该步骤将cDNA & gDNA片段化,同时进行末端修复及dA尾添加。
1. 将表3中各试剂解冻后,颠倒混匀,置于冰上备用。
2. 于冰上配制表3反应体系。
表3 DNA片段化/末端修复/dA尾添加 PCR反应体系
|
名称 |
体积(μL) |
名称 |
体积(μL) |
|
2nd Strand cDNA |
35 |
gDNA |
35 |
|
Smearase Buffer |
10 |
Smearase Buffer |
10 |
|
Smearase Enzyme Mix |
5 |
Smearase Enzyme Mix |
5 |
|
RNase-free H2O |
10 |
DNA Extra-working Buffer |
10 |
|
Total |
60 |
Total |
60 |
3. 使用移液器轻轻吹打或低速振荡混匀,并短暂离心将反应液离心至管底。
4. 将上述PCR管置于PCR仪,设置表4所示反应程序,进行DNA片段化,末端修复及dA尾添加反应。
表4 DNA片段化/末端修复/dA尾添加 PCR反应程序
|
温度 |
时间 |
|
热盖105°C |
on |
|
4°C |
1 min |
|
30 °C |
5–20 min** |
|
72 °C |
20 min |
|
4°C |
Hold |
【注】:*DNA片段化过程为有效控制片段化效果,避免过度酶切,反应程序可预先设置4°C,待模块温度降至4°C时,将PCR管放入PCR仪即可。**对于完整的基因组DNA,酶切时间参考表5。
表5 片段化时间选择表
|
插入片段主峰大小 |
片段化时间 |
|
300~500 bp |
5 min |
|
250 bp |
10 min |
|
200 bp |
15 min |
|
150 bp |
20~30 min |
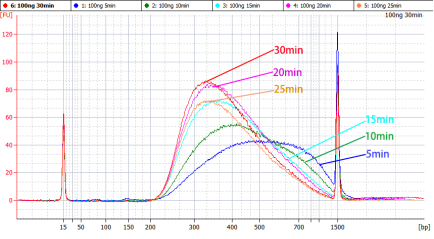
图2 不同片段化条件下的文库峰形参考
Step 2 接头连接(Adapter Ligation)
该步骤可在末端修复和dA尾添加的产物末端,连接特定的Illumina®或者MGI®接头。
1. 参考注意事项二中的表1,根据Input DNA,稀释Adapter至合适浓度。
2. 将表6中各试剂解冻后颠倒混匀,置于冰上备用。
3. 于Step 1步骤结束后的PCR管中继续配制表6所示反应体系。
表6 Adapter Ligation体系
|
名称 |
体积(μL) |
|
dA-tailed DNA |
60 |
|
Ligation Enhancer |
30* |
|
Novel T4 DNA Ligase |
5 |
|
DNA Adapter |
5** |
|
Total |
100 |
【注】:*Ligation Enhancer使用前请上下颠倒、振荡,充分混匀并瞬时离心后使用。
**本公司Illumina接头原始浓度为15 μM, 请根据注意事项二表1提示,根据投入量对接头进行稀释,使接头添加体积固定为5 μL。
4. 使用移液器轻轻吹打混匀,并短暂离心将反应液收集至管底。
5. 将PCR管置于PCR仪中,设置表7所示反应程序,进行接头连接反应:
表7 Adapter Ligation反应程序
|
温度 |
时间 |
|
热盖 |
Off |
|
20°C |
15 min |
|
4°C |
Hold |
Step 3连接产物纯化(Post Ligation Clean Up)
1. 准备工作:将Hieff NGS® DNA Selection Beads磁珠由冰箱中取出,室温平衡至少30 min。配制80%乙醇。
2. 涡旋振荡或充分颠倒磁珠以保证充分混匀。
3. 吸取45 μL Hieff NGS® DNA Selection Beads(0.45×,Beads:DNA=0.45:1)至Adapter Ligation产物中,涡旋或吹打混匀,室温孵育5 min。
4. 将PCR管短暂离心并置于磁力架中分离磁珠和液体,待溶液澄清后(约5 min),小心移除上清。
5. 保持PCR管始终置于磁力架中,加入200 μL新鲜配制的80%乙醇漂洗磁珠,室温孵育30 sec后,小心移除上清。
6. 重复步骤5,总计漂洗两次。
7. 保持PCR管始终置于磁力架中,用10 μL移液器吸干净残留液体,开盖空气干燥磁珠至刚刚出现龟裂(不超过5 min)。
8. 将PCR管从磁力架中取出,加入21 μL ddH2O,涡旋振荡或使用移液器轻轻吹打至充分混匀,室温静置5 min。将PCR管短暂离心并置于磁力架中静置,待溶液澄清后(约3 min),小心移取20 μL上清至新PCR管中,进行PCR扩增。
Step 4 文库扩增(Library Amplification)
该步骤将对纯化后的接头连接产物进行PCR扩增富集。
1. 将表8中的试剂解冻后颠倒混匀,置于冰上备用。
2. 于无菌PCR管中配制表8所示反应体系。
表8 短接头连接产物PCR反应体系(Illumina扩增体系)
|
组分名称 |
体积(μL) |
|
2× Ultima HF Amplification Mix |
25 |
|
Universal Primer/ i5 Primer* |
2.5 |
|
Index Primer/ i7 Primer* |
2.5 |
|
Adapter Ligated DNA |
20 |
|
Total |
50 |
【注】:*使用的是无Index的接头,俗称短接头(小Y接头),请使用短接头试剂(Cat#12412~Cat#12413)中配备的Index primer进行扩增。
3. 使用移液器轻轻吹打或振荡混匀,并短暂离心将反应液收集至管底。
4. 将PCR管置于PCR仪中,设置表9示反应程序,进行PCR扩增。
表9 PCR扩增反应程序
|
温度 |
时间 |
循环数 |
|
98°C |
1 min |
1 |
|
98°C |
10 sec |
参照注意事项三,文库扩增 |
|
60°C |
30 sec |
|
|
72°C |
30 sec |
|
|
72°C |
1 min |
1 |
|
4°C |
Hold |
– |
Step 5 扩增产物磁珠纯化(Post Amplification Clean Up)
1. 准备工作:将Hieff NGS® DNA Selection Beads磁珠由冰箱中取出,室温平衡至少30 min。配制80%乙醇。
2. 涡旋振荡或充分颠倒磁珠以保证充分混匀。
3. 吸取45 μL Hieff NGS® DNA Selection Beads(0.9×,Beads:DNA=0.9:1)至Adapter Ligation产物中,涡旋或吹打混匀,室温孵育5 min。
4. 将PCR管短暂离心并置于磁力架中分离磁珠和液体,待溶液澄清后(约5 min),小心移除上清。
5. 保持PCR管始终置于磁力架中,加入200 μL新鲜配制的80%乙醇漂洗磁珠,室温孵育30 sec后,小心移除上清。
6. 重复步骤5,总计漂洗两次。
7. 保持PCR管始终置于磁力架中,用10 μL移液器吸干净残留液体,开盖空气干燥磁珠至刚刚出现龟裂(不超过5 min)。
8. 将PCR管从磁力架中取出,加入32 μL ddH2O,涡旋振荡或使用移液器轻轻吹打至充分混匀,室温静置5 min。将PCR管短暂离心并置于磁力架中静置,待溶液澄清后(约3 min),小心移取30 μL上清至新PCR管中,进行文库定量、质检。
Step 6 文库质量控制
通常情况下,构建好的文库可通过浓度检测和长度分布检测来进行质量评价,具体请参见注意事项五。
HB220810
Hieff NGS® OnePot cDNA & gDNA Library Prep Kit是针对Illumina®或者MGI®测序平台专业开发设计的新一代酶切法建库试剂盒。与传统的建库法比较,本品采用高质量的片段化酶,摆脱了繁琐的超声过程,同时简化了操作流程,将片段化模块与末端修复模块合二为一,极大的降低了建库的时间和成本。本试剂盒具有优秀的文库转化率,可应用于常规动植物基因组、微生物基因组等样本,同时能兼容cfDNA样本的建库。该试剂盒使用了最新优化的连接酶,改善了接头连接时的片段自连现象,同时替换了新型高保真酶,进一步提升了扩增的均一性和保真性。
Ø 适用500 pg-1 μg的基因组DNA、全长cDNA(衔接Hieff NGS® ds-cDNA Synthesis Kit全长cDNA合成试剂盒(Yeasen Cat#13488))等样本;
Ø 高质量片段化酶,可随机切割双链DNA,酶切片段无偏好性;
Ø 片段化、末端修复/加A一步完成;
Ø 强扩增效率的高保真酶,显著提高文库质量及产量;
Ø 适用于cfDNA样本;
Ø 严格的批次性能与稳定性质控;
产品组分
|
组分编号 组分名称 |
13502ES24 |
13502ES96 |
||
|
13502-A |
 |
Smearase Buffer |
240 μL |
960 μL |
|
13502-B |
 |
DNA Extra-working Buffer |
240 μL |
960 μL |
|
13502-C |
 |
Smearase Enzyme Mix |
120 μL |
480 μL |
|
13502-D |
 |
Ligation Enhancer |
720 μL |
2×1440 μL |
|
13502-E |
 |
Novel T4 DNA Ligase |
120 μL |
480 μL |
|
13502-F |
 |
2× Ultima HF Amplification Mix |
600 μL |
2×1200 μL |
运输与保存方法
干冰运输。-20℃保存。有效期1年。
注意事项
一、关于操作
1. 为了您的安全和健康,请穿实验服并戴一次性手套操作。
2. 请于使用前将试剂盒各组分置于室温解冻。解冻后上下颠倒数次充分混匀,短暂离心后置于冰上待用。
3. 配制各步骤反应液时推荐使用移液器吹打混匀或轻轻振荡,剧烈振荡可能会造成文库产出下降。
4. 为避免样品交叉污染,推荐使用带滤芯的枪头,吸取不同样品时请更换枪头。
5. 推荐在带热盖的PCR仪中进行各步骤反应,使用前应预热PCR仪至反应温度附近。
6. PCR产物因操作不当极容易产生气溶胶污染,进而影响实验结果准确性。推荐将PCR反应体系配制区和PCR产物纯化检测区进行强制性的物理隔离;使用专用的移液器等设备;并定时对各实验区域进行清洁(使用0.5%次氯酸钠或10%漂白剂进行擦拭清理),以保证实验环境的洁净度。
7. 本产品仅作科研用途!
二、关于接头连接(Adapter Ligation)
Illumina接头:
1. 本公司可提供短接头(也称为小Y接头、不完整接头)试剂盒,客户可根据实验需求进行选择。
目前有双端 384 种 Index Primers: Hieff NGS® 384 CDI Primer for Illumina® , Set 1~Set 2 (Cat#12412~Cat#12413)。
2. 我们建议选用高质量的商业化接头。如客户使用自制接头,请委托具有NGS引物合成经验的公司,并备注需进行严格的防污染控制。此外,进行接头退火操作时,请在超净台完成。每次只操作一种接头,防止交叉污染。
3. 使用接头时,请提前将接头取出放在4°C或冰盒上解冻;在室温操作时,实验室温度最好不要超过25°C,防止接头解链。
4. 建库过程中,接头浓度过高或过低都会导致建库成功率变低。本试剂盒操作方案中,所加入的接头体积固定为5 μL,请根据初始的DNA或者RNA投入量,参考表1对接头进行稀释。本公司接头原始浓度均为15 μM,接头稀释液请选择0.1×TE buffer,稀释过的接头可在4°C保存48小时。
表1 Input Total RNA/DNA量与接头使用浓度推荐表
|
Input Total RNA(参照Cat#13488 RNA投入量) |
Adapter stock concentration |
Input Total DNA |
Adapter stock concentration |
|
≥10 ng |
15 μM |
1μg~200 ng |
15 μM |
|
<10 ng |
3 μM |
100 ng |
10 μM |
|
50 ng |
5 μM |
||
|
10 ng |
3 μM |
||
|
5 ng |
1μM |
||
|
≤1ng |
0.5μM |
三、关于文库扩增(Library Amplification)
文库扩增步骤需要严格控制扩增循环数。循环数不足,将导致文库产量低;循环数过多,又将导致文库偏好性增加、重复度增加、嵌合产物增加、扩增突变积累等多种不良后果。表2列举了使用本试剂盒,获得1mg文库的推荐循环数。
表 2 Input Total RNA或者DNA量与扩增循环数推荐表*
|
Input Total RNA |
Number of cycles |
Input Total DNA |
Number of cycles |
|
<1 ng |
10~12 |
<1 ng |
14~16 |
|
1ng |
9~10 |
1 ng |
13~14 |
|
10 ng |
6~7 |
5 ng |
10~11 |
|
50ng |
4~5 |
10 ng |
9~10 |
|
100~1000 ng |
4 |
50 ng |
7~8 |
|
100 ng |
6~7 |
||
|
200 ng |
5~6 |
【注】:*由于文库产量不仅与投入量和扩增循环数相关,样本质量等都会影响产量。建库过程中请根据实际情况综合考虑,选择最合适的建库条件。
四、DNA磁珠纯化与分选(Bead-based Clean Up and Size Selection)
1. 建库过程中有多个步骤需要使用DNA纯化磁珠,我们推荐使用Hieff NGS® DNA Selection Beads (Yeasen Cat#12601)或AMPure® XP磁珠(Beckman Cat#A63880)进行DNA纯化和分选。
2. 磁珠使用前应先平衡至室温,否则会导致得率下降、分选效果不佳。
3. 磁珠每次使用前都应充分振荡混匀或使用移液器上下吹打充分混匀。
4. 转移上清时,请勿吸取磁珠,即使微量残留都将影响后续文库质量。
5. 磁珠洗涤使用的80%乙醇应现用现配,否则将影响回收效率。
6. 产物洗脱前应将磁珠置于室温干燥。干燥不充分容易造成无水乙醇残留影响后续反应;过分干燥又会导致磁珠开裂进而降低纯化得率。通常情况下,室温干燥3-5 min足以让磁珠充分干燥。
7. DNA纯化或长度分选产物如需保存,可使用0.1×TE Buffer洗脱,产物于4°C可保存2天,-20°C可保存1个月。
五、关于文库质检(Library Quality Analysis)
1. 通常情况下,构建好的文库可通过长度分布检测和浓度检测来进行质量评价。
2. 文库浓度检测可使用:基于双链DNA荧光染料的方法,如Qubit®、PicoGreen®等;基于qPCR绝对定量的方法。
3. 推荐使用qPCR方法进行文库浓度检测:Qubit®等基于双链DNA荧光染料的浓度测定方法时,无法有效区分单端连接Adapter的产物、两端均未连接Adapter的产物及其他不完整双链结构产物;qPCR绝对定量基于PCR扩增原理,仅定量样品中两端Adapter完整的文库(即可测序的文库),可排除单端或双端都不连接Adapter的不可测序文库的干扰。
4. 文库浓度检测不可使用:基于光谱检测的方法,如NanoDrop®等。
5. 文库长度分布检测,可通过Agilent Bioanalyzer 2100等基于毛细管电泳或微控流原理的设备进行检测。
六、自备材料(Other Material)
1. DNA纯化磁珠:Hieff NGS® DNA Selection Beads(Yeasen Cat#12601)或AMPure® XP Beads(A63880)或其他等效产品。
2. Adapters:含Index的长接头(Yeasen Cat#12615~12618)或者无Index的短接头试剂盒(Yeasen Cat#12611~12614)。
3. 文库质检:Agilent 2100 Bioanalyzer DNA 1000 Chip/ High Sensitivity Chip或其他等效产品;文库定量试剂。
4. 其他材料:无水乙醇、无菌超纯水、低吸附枪头、PCR管、磁力架、PCR仪等。
建库流程图
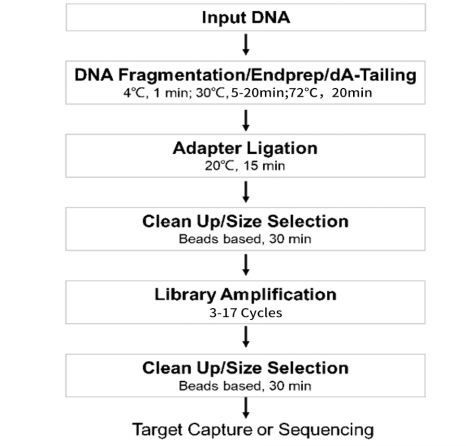
图1 DNA建库操作流程
使用方法
Step 1 cDNA & gDNA片段化/末端修复/dA尾添加(DNA Fragment/End Preparation/dA-Tailing)
该步骤将cDNA & gDNA片段化,同时进行末端修复及dA尾添加。
1. 将表3中各试剂解冻后,颠倒混匀,置于冰上备用。
2. 于冰上配制表3反应体系。
表3 DNA片段化/末端修复/dA尾添加 PCR反应体系
|
名称 |
体积(μL) |
名称 |
体积(μL) |
|
2nd Strand cDNA |
35 |
gDNA |
35 |
|
Smearase Buffer |
10 |
Smearase Buffer |
10 |
|
Smearase Enzyme Mix |
5 |
Smearase Enzyme Mix |
5 |
|
RNase-free H2O |
10 |
DNA Extra-working Buffer |
10 |
|
Total |
60 |
Total |
60 |
3. 使用移液器轻轻吹打或低速振荡混匀,并短暂离心将反应液离心至管底。
4. 将上述PCR管置于PCR仪,设置表4所示反应程序,进行DNA片段化,末端修复及dA尾添加反应。
表4 DNA片段化/末端修复/dA尾添加 PCR反应程序
|
温度 |
时间 |
|
热盖105°C |
on |
|
4°C |
1 min |
|
30 °C |
5–20 min** |
|
72 °C |
20 min |
|
4°C |
Hold |
【注】:*DNA片段化过程为有效控制片段化效果,避免过度酶切,反应程序可预先设置4°C,待模块温度降至4°C时,将PCR管放入PCR仪即可。**对于完整的基因组DNA,酶切时间参考表5。
表5 片段化时间选择表
|
插入片段主峰大小 |
片段化时间 |
|
300~500 bp |
5 min |
|
250 bp |
10 min |
|
200 bp |
15 min |
|
150 bp |
20~30 min |
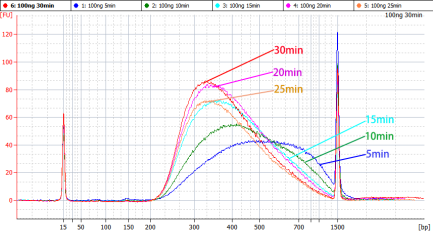
图2 不同片段化条件下的文库峰形参考
Step 2 接头连接(Adapter Ligation)
该步骤可在末端修复和dA尾添加的产物末端,连接特定的Illumina®或者MGI®接头。
1. 参考注意事项二中的表1,根据Input DNA,稀释Adapter至合适浓度。
2. 将表6中各试剂解冻后颠倒混匀,置于冰上备用。
3. 于Step 1步骤结束后的PCR管中继续配制表6所示反应体系。
表6 Adapter Ligation体系
|
名称 |
体积(μL) |
|
dA-tailed DNA |
60 |
|
Ligation Enhancer |
30* |
|
Novel T4 DNA Ligase |
5 |
|
DNA Adapter |
5** |
|
Total |
100 |
【注】:*Ligation Enhancer使用前请上下颠倒、振荡,充分混匀并瞬时离心后使用。
**本公司Illumina接头原始浓度为15 μM, 请根据注意事项二表1提示,根据投入量对接头进行稀释,使接头添加体积固定为5 μL。
4. 使用移液器轻轻吹打混匀,并短暂离心将反应液收集至管底。
5. 将PCR管置于PCR仪中,设置表7所示反应程序,进行接头连接反应:
表7 Adapter Ligation反应程序
|
温度 |
时间 |
|
热盖 |
Off |
|
20°C |
15 min |
|
4°C |
Hold |
Step 3连接产物纯化(Post Ligation Clean Up)
1. 准备工作:将Hieff NGS® DNA Selection Beads磁珠由冰箱中取出,室温平衡至少30 min。配制80%乙醇。
2. 涡旋振荡或充分颠倒磁珠以保证充分混匀。
3. 吸取45 μL Hieff NGS® DNA Selection Beads(0.45×,Beads:DNA=0.45:1)至Adapter Ligation产物中,涡旋或吹打混匀,室温孵育5 min。
4. 将PCR管短暂离心并置于磁力架中分离磁珠和液体,待溶液澄清后(约5 min),小心移除上清。
5. 保持PCR管始终置于磁力架中,加入200 μL新鲜配制的80%乙醇漂洗磁珠,室温孵育30 sec后,小心移除上清。
6. 重复步骤5,总计漂洗两次。
7. 保持PCR管始终置于磁力架中,用10 μL移液器吸干净残留液体,开盖空气干燥磁珠至刚刚出现龟裂(不超过5 min)。
8. 将PCR管从磁力架中取出,加入21 μL ddH2O,涡旋振荡或使用移液器轻轻吹打至充分混匀,室温静置5 min。将PCR管短暂离心并置于磁力架中静置,待溶液澄清后(约3 min),小心移取20 μL上清至新PCR管中,进行PCR扩增。
Step 4 文库扩增(Library Amplification)
该步骤将对纯化后的接头连接产物进行PCR扩增富集。
1. 将表8中的试剂解冻后颠倒混匀,置于冰上备用。
2. 于无菌PCR管中配制表8所示反应体系。
表8 短接头连接产物PCR反应体系(Illumina扩增体系)
|
组分名称 |
体积(μL) |
|
2× Ultima HF Amplification Mix |
25 |
|
Universal Primer/ i5 Primer* |
2.5 |
|
Index Primer/ i7 Primer* |
2.5 |
|
Adapter Ligated DNA |
20 |
|
Total |
50 |
【注】:*使用的是无Index的接头,俗称短接头(小Y接头),请使用短接头试剂(Cat#12412~Cat#12413)中配备的Index primer进行扩增。
3. 使用移液器轻轻吹打或振荡混匀,并短暂离心将反应液收集至管底。
4. 将PCR管置于PCR仪中,设置表9示反应程序,进行PCR扩增。
表9 PCR扩增反应程序
|
温度 |
时间 |
循环数 |
|
98°C |
1 min |
1 |
|
98°C |
10 sec |
参照注意事项三,文库扩增 |
|
60°C |
30 sec |
|
|
72°C |
30 sec |
|
|
72°C |
1 min |
1 |
|
4°C |
Hold |
– |
Step 5 扩增产物磁珠纯化(Post Amplification Clean Up)
1. 准备工作:将Hieff NGS® DNA Selection Beads磁珠由冰箱中取出,室温平衡至少30 min。配制80%乙醇。
2. 涡旋振荡或充分颠倒磁珠以保证充分混匀。
3. 吸取45 μL Hieff NGS® DNA Selection Beads(0.9×,Beads:DNA=0.9:1)至Adapter Ligation产物中,涡旋或吹打混匀,室温孵育5 min。
4. 将PCR管短暂离心并置于磁力架中分离磁珠和液体,待溶液澄清后(约5 min),小心移除上清。
5. 保持PCR管始终置于磁力架中,加入200 μL新鲜配制的80%乙醇漂洗磁珠,室温孵育30 sec后,小心移除上清。
6. 重复步骤5,总计漂洗两次。
7. 保持PCR管始终置于磁力架中,用10 μL移液器吸干净残留液体,开盖空气干燥磁珠至刚刚出现龟裂(不超过5 min)。
8. 将PCR管从磁力架中取出,加入32 μL ddH2O,涡旋振荡或使用移液器轻轻吹打至充分混匀,室温静置5 min。将PCR管短暂离心并置于磁力架中静置,待溶液澄清后(约3 min),小心移取30 μL上清至新PCR管中,进行文库定量、质检。
Step 6 文库质量控制
通常情况下,构建好的文库可通过浓度检测和长度分布检测来进行质量评价,具体请参见注意事项五。
HB220810

暂无内容

暂无内容
